Inconsistent Data Sets: Normalizing CMS Quality Data Files for Analysis
In the last post, we discussed the five file formats encountered by Freedman Analytical Engine (FAE) as it processes Centers for Medicare and Medicaid Services (CMS) quality data. They require FAE to standardize these files to create one merged CSV. This merged file will most closely resemble a Vertical ID file. This post will focus on the two most complex file structures, Transposed and Horizontal.
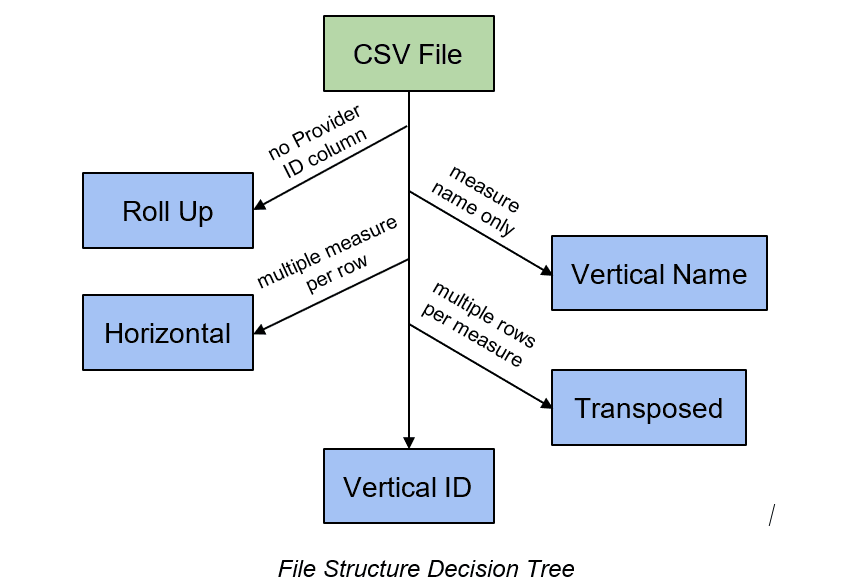
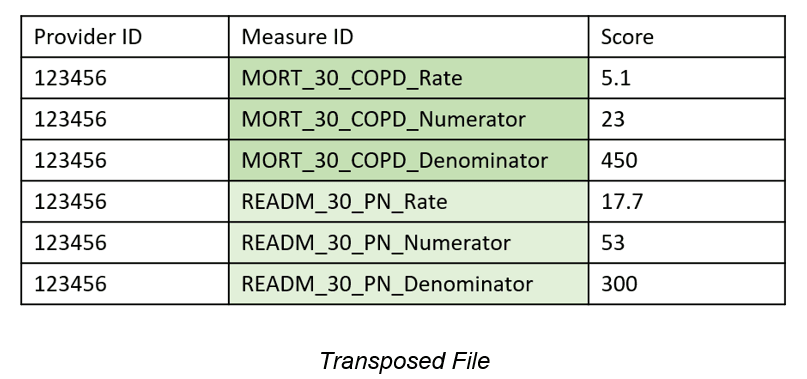
As you can see in the decision tree above, the last two possible files are Vertical ID and Transposed. The only difference is that, in Transposed files, there are multiple rows relating to the same measure, so they have a consistent prefix. To identify Transposed files, FAE looks for repeated prefixes. These files are not automatically marked as transposed because, on occasion, unique measures in a Vertical ID file appear to have matching prefixes. A programmer or an analyst can look at these potentially Transposed files to decide. By allowing for augmentation rather than automation in this step, we catch exceptions and categorize appropriately.
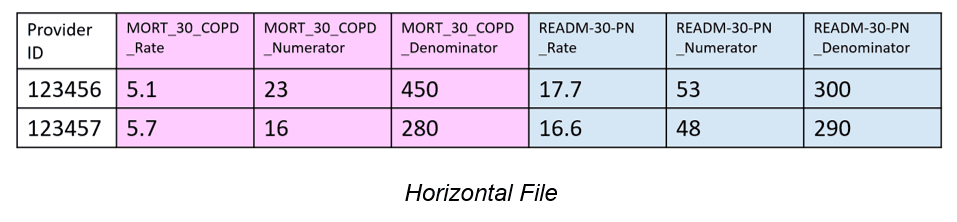
Identifying Horizontal files can be done automatically by finding multiple measure identifier column headers. After the identification step, each Horizontal file row needs to be broken into multiple rows. In the case of the Horizontal file depiction we’ve seen, also seen below, MORT_30 and READM_30_PN should have their own rows. They should each be measures in a Measure ID column. Rate, Numerator, and Denominator should be their own columns because they are attributes of the measures. By picking out groups or chunks of measures (those related to MORT_30 and those related to READM_30_PN), we can create a normalized file with rows for each measure. To the human eye, separating MORT_30 and READM_30_PN files is clear, but there are instances where the distinction doesn’t follow a clear, programmable pattern for FAE to use. We use the measure dictionary to determine where one measure ends and another starts, thus grouping the columns.
Though dealing with these varying file structures makes the process more complex, FAE’s method requires little interaction from programmers or analysts. FAE both identifies and normalizes these formats into a CSV that can be analyzed as one dataset.
In the next post, we will discuss a summary of the ETL normalization of CMS Quality Data.
